UNDER DEVELOPMENT
Creating software for specific biological fields in the academic research setting requires a unique set of skills. In this blog I’ve summarized these skill sets into three areas.
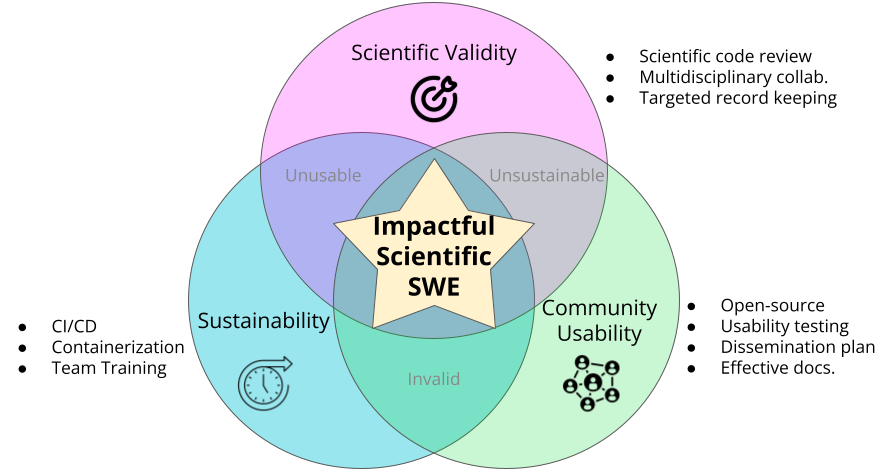
- Scientific Validity - Software data handling and methods need to be congruent and effective for the scientific questions at hand. This requires biomedical knowledge to enforce. Scientific validity is ensured through the use of effective multidisciplinary collaboration, robust scientific code review through pull requests, and robust record keeping of quality controls and sanity checks data analyses.
- Multidisciplinary communication is necessary
- Clear communication is necessary
- Scientific code review is necessary
- Community Usability - Scientific software needs to be usable by the community in order to impact research. Usability is fundamentally increased through making work open source and transparent with a permissive license for reuse. Principles of usability and effective documentation are required so that the intended audience of researchers are able to find the information they need easily to apply the software to their own work.
- Power of three - you mostly need just three testers to see what the problems are
- Training as a usability tool - Most academic software isn’t documented enough for others to understand or use.
- Sustainability - Scientific software cannot be impactful if it is not lasting. In an academic research setting, software sustainability can be particularly tricky if not properly funded or if the engineer doesn’t require the proper skill set. Software is made more sustainable through unit testing and continuous integration and continuous deployment methods which ensure that new changes to the code do not break current features. Containerization is a critical tool for software sustainability as irreconcilable software dependencies can easily derail a biological data analysis before it has even begun. Lastly a technique for sustainability is to make sure others on the team are properly trained in these techniques.
- CI/CD - Using this will help you test your software when you are a small or large team.
- Containerization - using this will help provide snapshots of computing environments you need
- Commensalistic symbiotic software - borrowing bigger, more robust software to be your back end, even if it doesn’t fully fit your needs. This can be helpful for maintenance burdens for small teams.
- Training as a software sustainability tool
R version 4.4.0 (2024-04-24)
Platform: x86_64-apple-darwin20
Running under: macOS 15.0.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] vctrs_0.6.5 httr_1.4.7 cli_3.6.3 knitr_1.49
[5] rlang_1.1.4 xfun_0.49 jsonlite_1.8.9 glue_1.8.0
[9] openssl_2.2.2 askpass_1.2.1 htmltools_0.5.8.1 hms_1.1.3
[13] fansi_1.0.6 rmarkdown_2.29 evaluate_1.0.1 tibble_3.2.1
[17] tzdb_0.4.0 fastmap_1.2.0 yaml_2.3.10 lifecycle_1.0.4
[21] compiler_4.4.0 ottrpal_1.2.1 fs_1.6.5 htmlwidgets_1.6.4
[25] pkgconfig_2.0.3 rstudioapi_0.17.1 digest_0.6.37 R6_2.5.1
[29] utf8_1.2.4 readr_2.1.5 pillar_1.9.0 magrittr_2.0.3
[33] tools_4.4.0 xml2_1.3.6
